TRAIN SET - Linear Model R^2 = 0.44; Polynomial Model R^2 = 0.62
VALID SET - Linear Model R^2 = 0.65; Polynomial Model R^2 = 0.41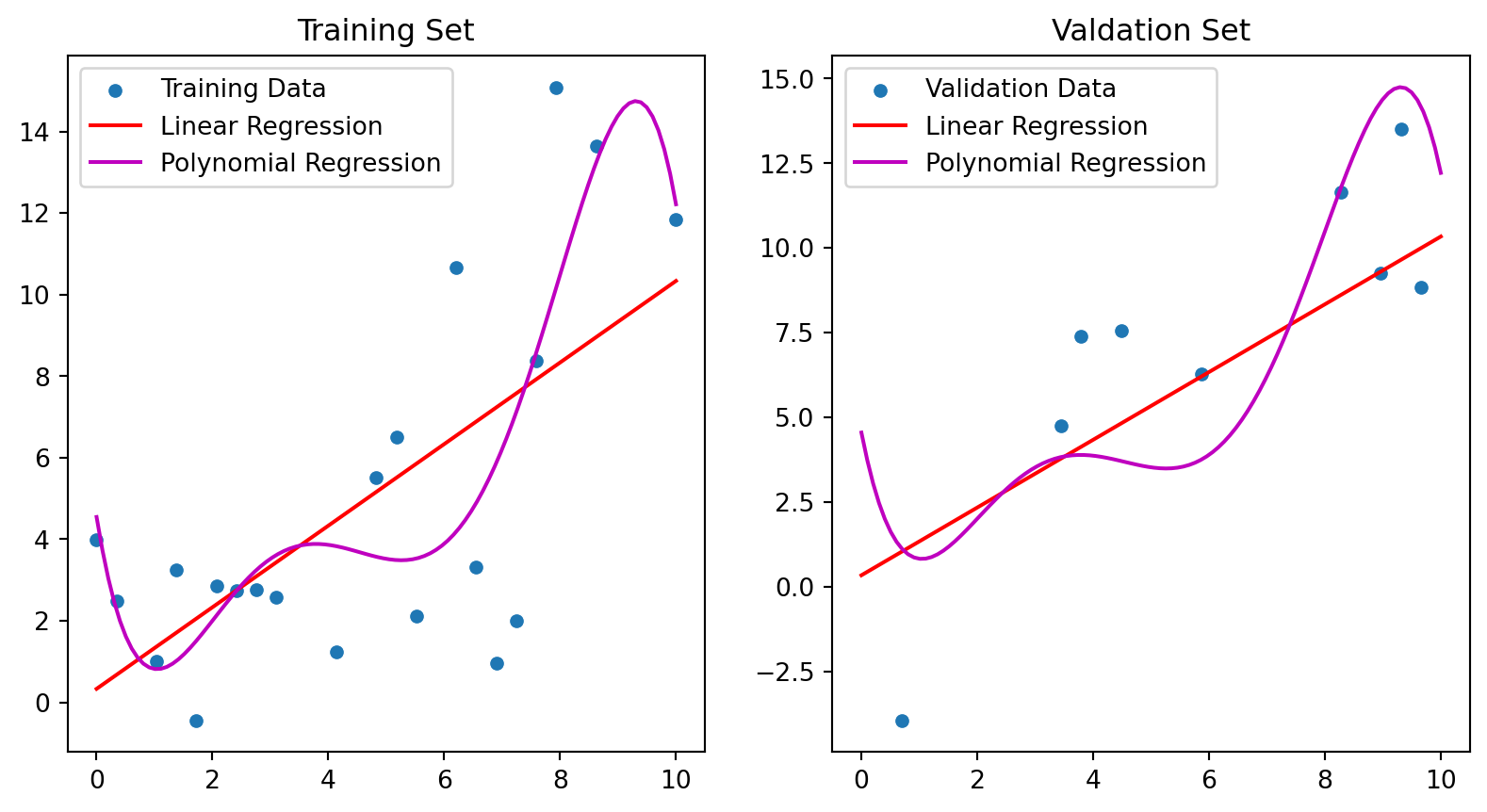
Evaluating Model Performance - Bootsrapping
TRAIN SET - Linear Model R^2 = 0.44; Polynomial Model R^2 = 0.62
VALID SET - Linear Model R^2 = 0.65; Polynomial Model R^2 = 0.41


n_boots = 100 # number of bootrap iterations
n_points = int(len(df) * 0.50) # sample size with replacement
boot_slopes = [] # store regressed line slopes
boot_intercepts = [] # store regressed line intercepts
plt.figure() # creates a figure that we can plot *multiple* times
linear_model = LinearRegression()
for _ in range(n_boots):
# sample the rows, same size, with replacement
sample_df = df.sample(n=n_points, replace=True)
# fit a linear regression
linear_model.fit(sample_df[['flipper_length_mm']], sample_df['body_mass_g'])
# append regressed coefficients
boot_intercepts.append(linear_model.intercept_)
boot_slopes.append(linear_model.coef_[0])
# plot a greyed out line of the prediction
y_pred_temp = linear_model.predict(sample_df[['flipper_length_mm']])
plt.plot(sample_df['flipper_length_mm'], y_pred_temp, color='grey', alpha=0.2)# add data points
plt.scatter(sample_df['flipper_length_mm'], sample_df['body_mass_g'])
plt.grid(True)
plt.xlabel('Flipper Length')
plt.ylabel('Body Mass')
plt.title(f'Bootrapping; Bootstrap interations={n_boots}; Sample Size:{n_points}')
plt.show();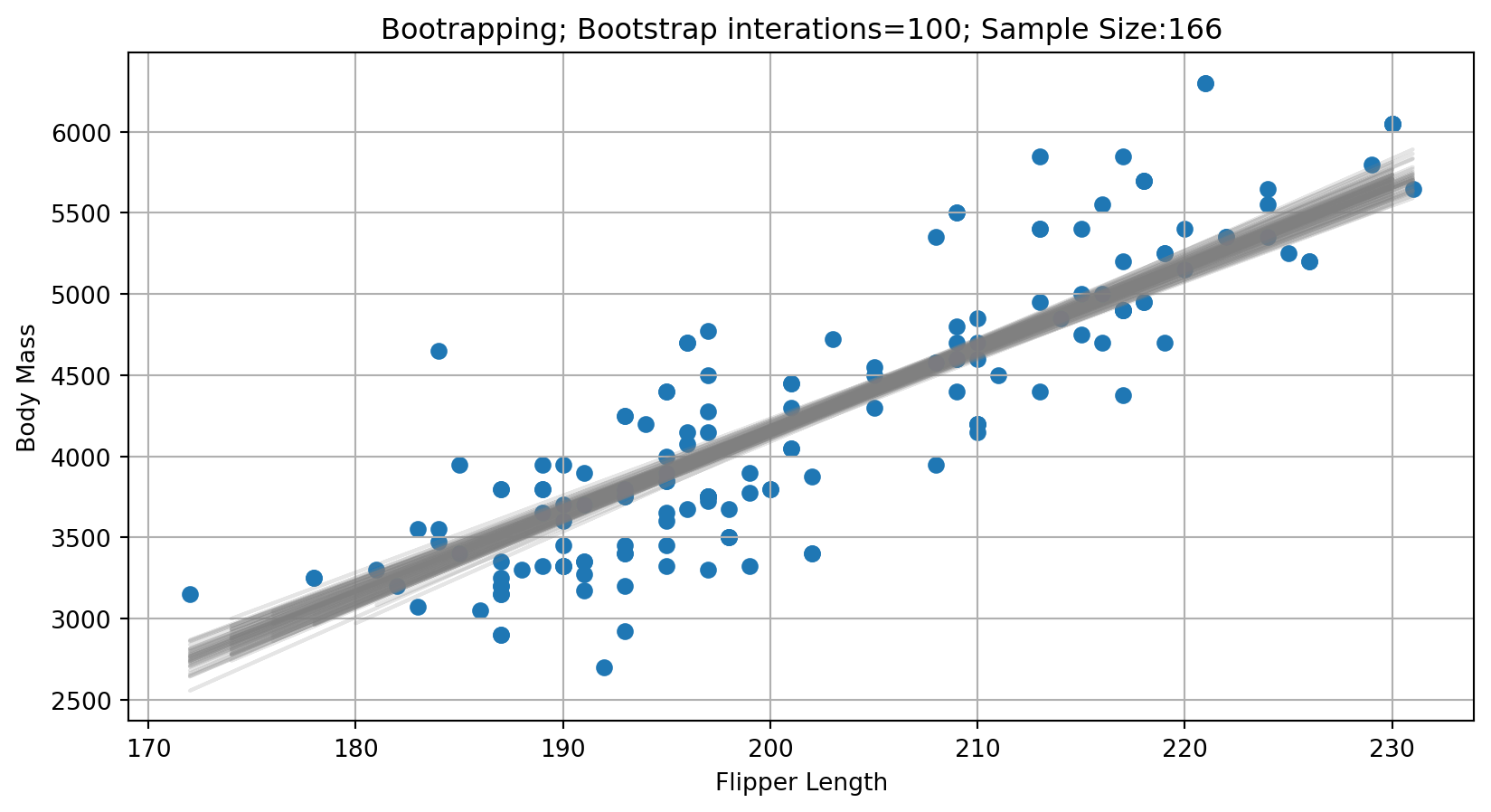
How consistent are the predictions for different samples?